VetGPT - RAG
- Overview
- RAG (Retrieval Augmented Generation)
- What’s needed?
- 1. Boilerplate FastAPI Project
- 2. Setup the database
- 3. Ingestion
- 4. Querying the model
- 5. Response
- Conclusion
- Resources
Overview
I love AI. Will it take over my job one day? Probably. Like most jobs, at some point something will come along that will do it better than I can. I’ll then either pivot, choose a different career or become a goose farmer. Who knows? But until then, I love how more efficient life and work has become. As a result, I’ve started delving into the technical side of AI (A bit late to the party but jumping on every trend until it becomes mainstream is a gamble. NFTs for example…)
My dog has recently had TPLO surgery - She’s doing fine but it provided some inspiration to create a RAG (Retrieval Augmented Generation) system that can answer questions about her surgery and recovery. This post will cover how I set up VetGPT.
This isn’t a complete project yet and I’ve got many more areas to cover including fine-tuning the model, improving the user interface, integrating with other systems, and eventually get this into the cloud (Which weirdly I know a lot more about than doing this locally).
The full repository can be found on here
Disclaimer: AI has helped with any problems faced rather than generating the solution. GenAI is great for generating boilerplate code and helping with issues, but I find the best way of learning is doing.
RAG (Retrieval Augmented Generation)
Tuning a model takes time, electricity and a lot of money. RAG is a way to use a pre-trained model and augment it with your own data. This allows you to get the benefits of a custom model without the cost of training one from scratch.
You essentially have a pre-trained model that can answer general questions, and then you provide it with additional context or data to improve its responses for specific topics. In this case, the topic is anything veterinary related.
Think about it like this: You have a doctor who knows a lot about the human body. You wouldn’t expect them to remember every single detail about every patient they’ve ever seen, right? Instead, they have access to medical records, research papers, and other resources that help them make informed decisions. RAG works in a similar way by providing the model with relevant information to enhance its understanding and responses.
What’s needed?
So doing some research, I needed the following:
- A pre-trained model that won’t destroy my computer: I chose Phi-3-mini, which is a smaller version of the Phi-3 model.
- A FastAPI project. Easy enough to setup.
- A fair bit of data to give to the model.
- A vector database to store the data and allow for quick retrieval. I used Qdrant - Easy enough to set up.
1. Boilerplate FastAPI Project
Boilerplate FastAPI project is easy enough to set up. An endpoint for querying the model. This AI Stuff is ready right?
main.py
@app.post("/q")
async def query(query: Query):
# Logic will come in here later on
2. Setup the database
Qdrant is a vector database that allows you to store and retrieve vectors efficiently. It’s fairly easy to use and setup.
db/qdrany.py
import os
from qdrant_client import QdrantClient
qdrant = QdrantClient(
host=os.getenv("QDRANT_HOST", "localhost"),
port=int(os.getenv("QDRANT_PORT", "6333")),
api_key=os.getenv("QDRANT_API_KEY", None),
https=os.getenv("QDRANT_HTTPS", "False").lower() == "true",
)
docker-compose.yml
services:
qdrant:
image: qdrant/qdrant:latest
container_name: qdrant
ports:
- "6333:6333"
restart: unless-stopped
Easy enough.
3. Ingestion
To keep things simple, I decided to use a list of PDFs with vetinary related information and ingest them into the vector database. Few key bits to note:
- SentenceTransformers: This is used to convert the text into vectors (embeddings) that can be stored in the vector database. A great article on how this all works can be found here.
- QdrantClient: This is used to interact with the Qdrant vector database.
The folder structure for the PDFs is as follows:
vet-gpt/
├── resources/
│ ├── procedures/
│ │ ├── tplo
│ │ │ ├── tplo.pdf
│ │ │ └── tplo2.pdf
└── main.py
The ingestion script will read the PDFs, tag the procedure from the folder structure, and then store the vectors in the Qdrant database.
ingest/ingest_procedures.py
import uuid
from dotenv import load_dotenv
from pypdf import PdfReader
from qdrant_client.http.models import Distance, VectorParams
from sentence_transformers import SentenceTransformer
load_dotenv()
model = SentenceTransformer("all-MiniLM-L6-v2")
import sys
from pathlib import Path
# Add the project root to the Python path to allow importing from sibling directories
current_dir = Path(__file__).parent
project_root = current_dir.parent.resolve()
sys.path.insert(0, str(project_root))
# Now we can import from the db module
from db.qdrant import qdrant
# Get the absolute path to the procedures directory
current_dir = Path(__file__).parent
procedures_dir = (current_dir / ".." / "resources" / "procedures").resolve()
# Check if collection exists
if not qdrant.collection_exists("procedures"):
print("Creating 'procedures' collection in Qdrant...")
qdrant.create_collection(
collection_name="procedures",
vectors_config=VectorParams(
size=384,
distance=Distance.COSINE,
)
)
else:
print("'procedures' collection already exists in Qdrant.")
# Iterate through each procedure directory
for procedure_dir in procedures_dir.iterdir():
if procedure_dir.is_dir():
procedure_name = procedure_dir.name
print(f"Found procedure: {procedure_name}")
# Iterate through files in the procedure directory
for file_path in procedure_dir.glob("**/*"):
if file_path.is_file():
# If file ends with .txt, we can skip it
if file_path.suffix.lower() == ".txt":
continue
print(f" - File: {file_path.relative_to(procedure_dir)}")
pdf_reader = PdfReader(str(file_path))
text = " ".join(page.extract_text() for page in pdf_reader.pages if page.extract_text())
chunks = [text[i:i + 500] for i in range(0, len(text), 500)]
for chunk in chunks:
embedding = model.encode(chunk).tolist()
qdrant.upsert(
collection_name="procedures",
points=[
{
"id": str(uuid.uuid4()),
"vector": embedding,
"payload": {
"text": chunk,
"procedure": procedure_name,
"file_name": file_path.name,
"source": "application/pdf"
}
}
]
)
Messy at the moment as I want to abstract this to work with other topics such as medications, nutrition, etc. And eventually I want to support other file types such as images, videos, etc.
Qdrant Dashboard

4. Querying the model
Now that we have the data in the vector database, we can query it using the pre-trained model. The query will take the user’s question, convert it into a vector using the same SentenceTransformer model, and then search the Qdrant database for the most relevant vectors. The retrieved vectors will be used to provide context to the pre-trained model, which will generate a response based on the question.
Now locally on my machine, responses take up to 20 seconds to generate. In a production environment, this would be much faster as the model would be hosted on a server with more resources. But for now, this works.
We feed the document chunks, and the prompt to the model.
main.py
from dotenv import load_dotenv
from fastapi import FastAPI
from pydantic import BaseModel
from sentence_transformers import SentenceTransformer
from llama_cpp import Llama
# Load environment variables from a .env file
load_dotenv()
# Import the Qdrant client singleton
from db.qdrant import qdrant
app = FastAPI()
print("[Sentence Transformer] Initializing...")
sentence_transformer = SentenceTransformer("all-MiniLM-L6-v2")
print("[Sentence Transformer] Initialized.")
print("[LLM] Initializing...")
LLM_MODEL_PATH = "./models/Phi-3-mini-128k-instruct.Q4_0.gguf"
llm = Llama(
model_path=LLM_MODEL_PATH,
n_ctx=4096,
n_gpu_layers=0, # Set to 0 for CPU-only inference. Set to -1 to offload all layers to GPU if available.
verbose=False
)
print(f"[LLM] Llama.cpp LLM Initialized from {LLM_MODEL_PATH}.")
class Query(BaseModel):
q: str
@app.post("/q")
async def query(query: Query):
query_embedding = sentence_transformer.encode(query.q).tolist()
search_results = qdrant.query_points(
collection_name="procedures",
query=query_embedding,
limit=5,
with_payload=True,
)
context_parts = []
sources = set()
for hit in search_results.points:
chunk_text = hit.payload.get("text", "N/A")
procedure_name = hit.payload.get("procedure", "N/A")
file_name = hit.payload.get("file_name", "N/A")
category = hit.payload.get("category", "N/A")
# Format context for the LLM, including metadata
context_parts.append(
f"--- Document Chunk ---\n"
f"Category: {category}\n" # Include category if you added it
f"Item: {procedure_name}\n"
f"Source File: {file_name}\n"
f"Content: {chunk_text}"
)
sources.add(f"{category or procedure_name} - {file_name}") # Better source tracking
context_string = "\n\n".join(context_parts)
system_message = (
"<|system|>"
"You are a helpful and knowledgeable veterinary AI assistant. Your goal is to provide accurate and concise answers based *only* on the provided veterinary information. "
"Each piece of information is clearly labeled with its 'Category', 'Item', and 'Source File'. If the information is insufficient to answer the question, say so and do not invent details. "
"Always cite the 'Source File' you use.<|end|>"
)
user_message = f"""<|user|>
Carefully read the 'Content' from the relevant documents below, then answer my question.
--- Veterinary Information ---
{context_string}
--- End of Veterinary Information ---
My Question: {query.q}<|end|>"""
prompt_template = system_message + user_message + "<|assistant|>"
print("[LLM] Sending prompt to LLM...")
output = llm(
prompt_template,
max_tokens=1000,
temperature=0.2,
stop=["<|endoftext|>", "<|end|>"],
)
print("[LLM] Sent prompt to LLM...")
llm_response_text = output["choices"][0]["text"].strip()
print(llm_response_text)
return {
"answer": llm_response_text,
"sources": list(sources)
}
5. Response
Model Response
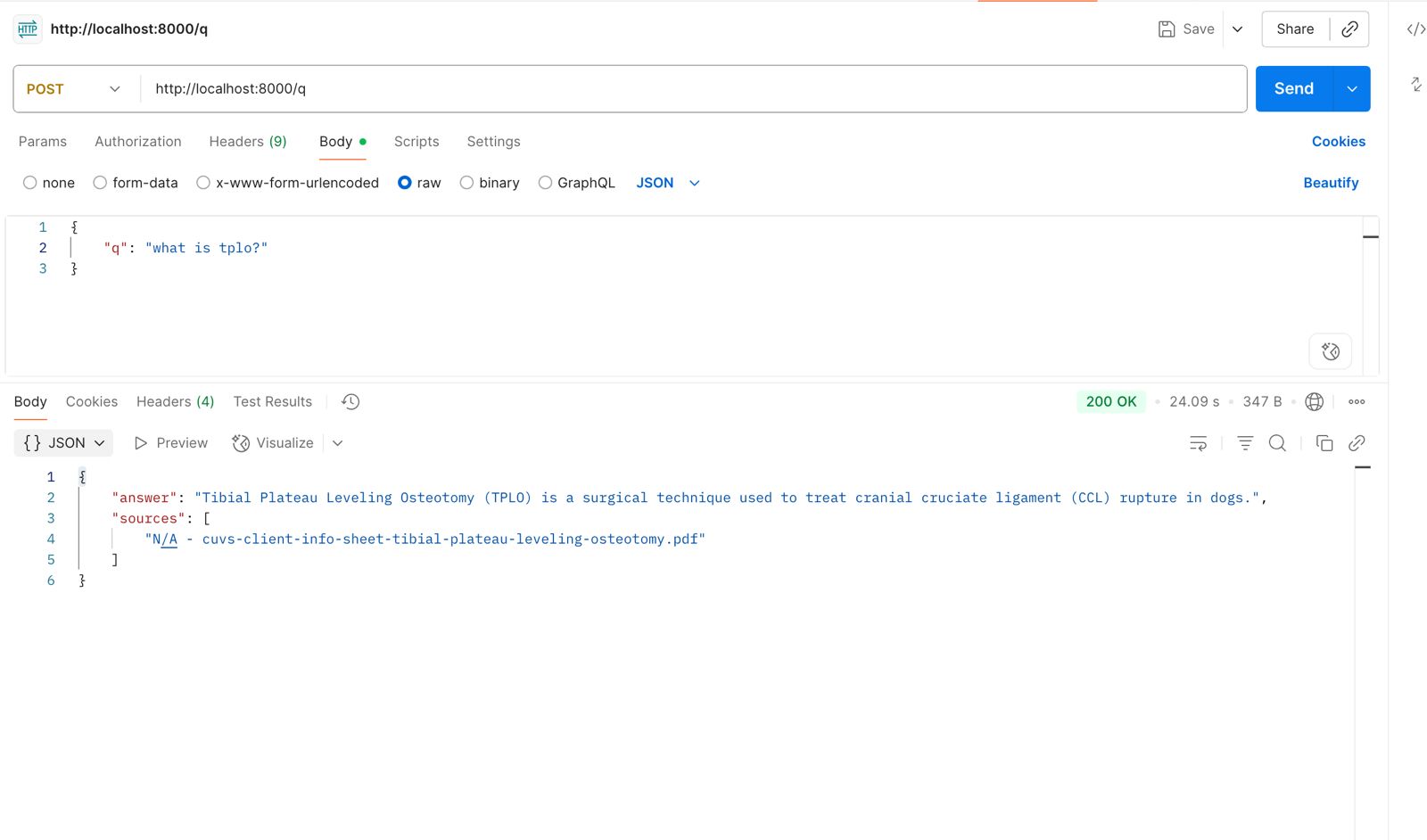
Hell yeah this is exciting. With information fed from simple PDFs, we can get a response that is relevant to the question asked. As more data is added, we can ask more complex question, get more detailed responses and eventually get this into a web interface that can be used by vets and pet owners alike.
Conclusion
So there’s many things I’ll be experimenting and tweaking with:
- Temperature: The responses are very specific. Maybe too specific. I might need to increase the temperature to get more varied responses.
- Max Tokens: The responses are limited to 1000 tokens which is fine for now but might need to be increased for more complex questions. Especially if we ask about multiple procedures or medications.
- Run the model on a home server w/ GPU: This will speed up the responses significantly. I have a HPE server but I need to get a GPU for it. This will help with fine-tuning the model as well.
- Fine-tuning the model: I want to fine-tune the model with more specific data and improve its responses
- AWS Bedrock + SageMaker: I want to explore using AWS Bedrock and SageMaker for more scalable and efficient model training and deployment
- Throw numbers at it: Average lab results, move into the field of processing lab values, highlighting abnormalities, and providing insights based on the data.
As I said, this is a simple concept project more than anything. But provides a good foundation to build on. AI is here to stay and it’s a fascinating field to be involved in.